R绘制漂亮的图一
Table of Content
普通plot绘图
绘制折线图
绘制折线图,并设置合适的坐标字体
par(mar=c(4.8,4.8,2,2))
plot(1:(n_egv-1), egrat, type='o', xlab='q', ylab='eigv. ratio', lwd=3, cex.axis = 1.6, cex.lab=1.8)
grid(col=1, lwd=2, lty=2)坐标轴标签旋转绘图
par(mar=c(6,6,2,2))
plot(1:nrow(dat_Tr_sort_inter), axes=F,
dat_Tr_sort_inter$total_trainhours[order_Tr],
type='o', xlab='', ylab='Total_hours', lwd=3, cex.axis = 1.6, cex.lab=1.8)
grid(col=1, lwd=2, lty=2)
axis(1, 1:nrow(dat_Tr_sort_inter),labels=dat_Tr_sort_inter$Enames[order_Tr],
las = 2)
axis(2)4 怎么添加图像的形状参数,有的曲线有形状,有的曲线没有形状,没有形状的曲线用-1表示
legend('topleft', c('真实曲线', '预测曲线', '拐点时间'), col=c(1,2,4), lty=1, pch=c(1,-1,-1))5. R绘制长度不同的向量的箱线图
Rt_list <- list(Rt_Pro, Rt_Hubei, Rt_Wuhan, Rt_Japan, Rt_Skorea, Rt_Sing, Rt_Italy)
boxplot(Rt_list, ylim=c(0,5.2), names = c('China', 'Hubei', 'Wuhan', 'Japan', 'SKorea', 'Singapore',
'Italy'))
abline(h=1, lty=3,col='green4')6. 添加没有边框的图例:利用bty=‘n’
plot(1: (nr-id_vec[1]+ 1-time), ratio7(dat[(id_vec[1]): nr,1], time),
xlim=xlim, ylim=ylim, col=1,
type='o', xlab='Day', ylab='ratio')
for(k in 2: nc){
lines(1: (nr-id_vec[k]+ 1-time), ratio7(dat[(id_vec[k]): nr,k], time), col=k)
points(1: (nr-id_vec[k]+ 1-time), ratio7(dat[(id_vec[k]): nr,k], time), col=k, pch=k)
}
abline(h =1 ,col=1, lty=2)
legend('topright', names(dat), col=1:nc, pch=1:nc,lty=1, bty='n')
2 漂亮美观强大的ggplot2绘图技术
Basic plots
1 绘制箱线图
datafun <- function(mat1, mat2, xname, methodname){
n1 <- dim(mat1)[1];
n2 <- dim(mat2)[1];
p <- dim(mat1)[2];
suml <- (n1 + n2)*p
data = as.data.frame(matrix(0, suml, 1))
for(i in 1:p){
data[((i - 1)*n1 + 1):(n1*i), 1] <- mat1[, i]
}
for(j in 1:p){
data[(n1*p + (j - 1)*n2 + 1):(n1*p + n2*j), 1] <- mat2[, j]
}
colnames(data) = "Value";
data$method = c(rep(methodname[1], n1*p), rep(methodname[2], n2*p))
data$snp_sparse = c(rep(xname, each = n1), rep(xname, each = n2))
return(data)
}
library("mvtnorm")## Warning: package 'mvtnorm' was built under R version 4.0.5library("ggplot2")
n1 <- 50;
n2 <- 100;
p <- 4;
mat1 <- rmvnorm(n1, mean = rep(0, p), sigma = diag(p), method = "chol");
mat2 <- rmvnorm(n1, mean = rep(0, p), sigma = diag(p), method = "chol");
xname = c("0.2", "0.3", "0.4", "0.5");
methodname = c("method1","method2");
dat <- datafun(mat1, mat2, xname, methodname)
# 按照x的值分组画箱线图,并设置坐标字体大小
ggplot(dat, aes(x=snp_sparse, y=Value, fill=method)) + geom_boxplot(aes(fill=method)) +
theme(axis.text.x = element_text(size = 12, angle=70, vjust=0.6)
,axis.text.y = element_text(size = 18),plot.title = element_text(hjust = 30,vjust=-40,size=14)
,legend.position="none") +ylim(c(0,2))## Warning: Removed 216 rows containing non-finite values (stat_boxplot).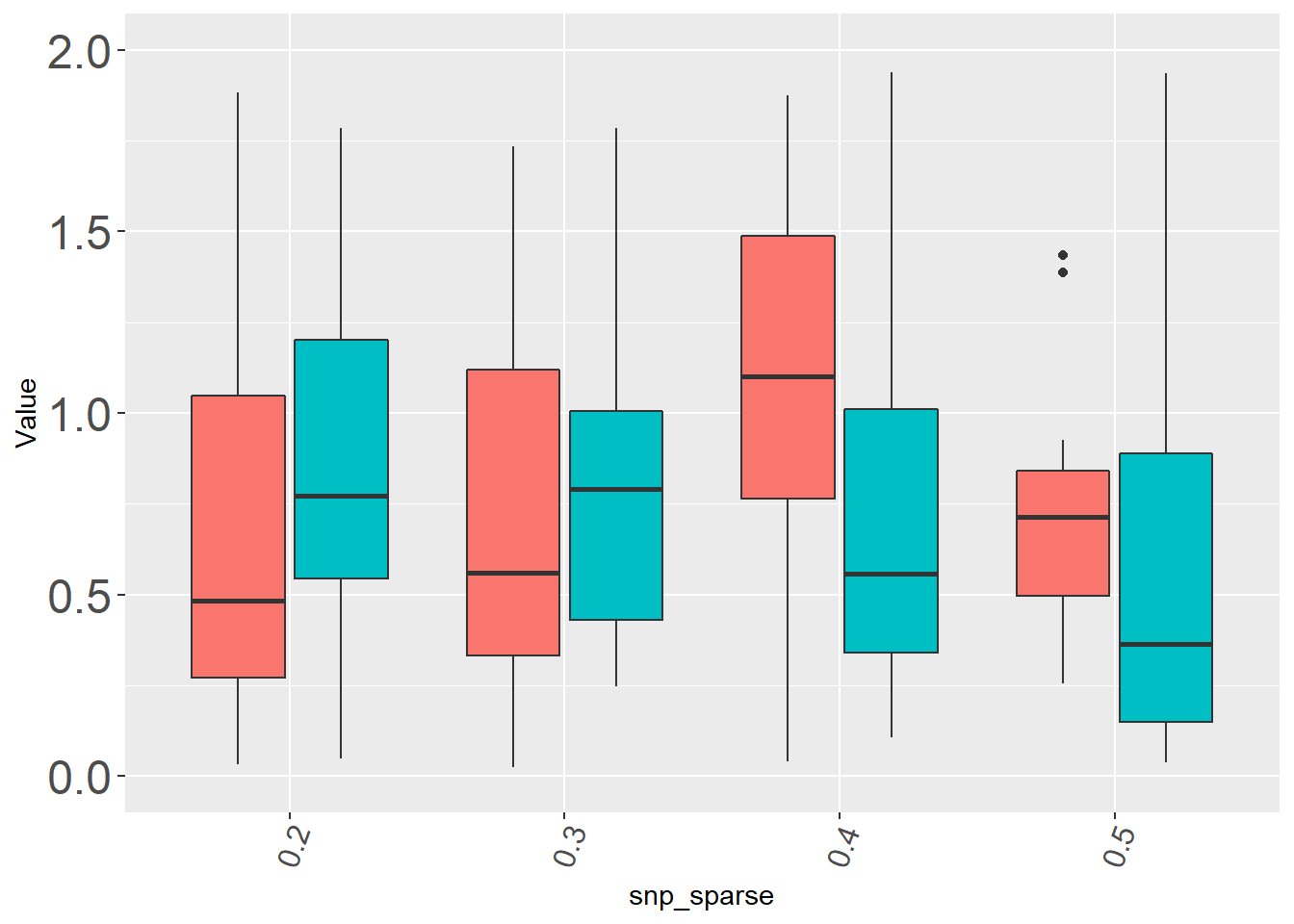
2 利用ggplot2画折线图
elboPlot <- function(elbo_seq){
library(ggplot2)
p1 <- ggplot(data=data.frame(ELBO= elbo_seq, iter=1:length(elbo_seq)),
aes(x=iter, y=ELBO))
p1 + geom_line(size=2) + ggtitle("VB algorithm") +
theme(plot.title = element_text(hjust = 0.5),
axis.text.x=element_text(size=14,color="red"),
axis.text.y=element_text(size=14, color="green"),
axis.title.x = element_text(size=18, color='black'),
axis.title.y = element_text(size=18, color='black'),
title= element_text(size=20, color='blue')
)
}
elboPlot(elbo_seq = 1:100)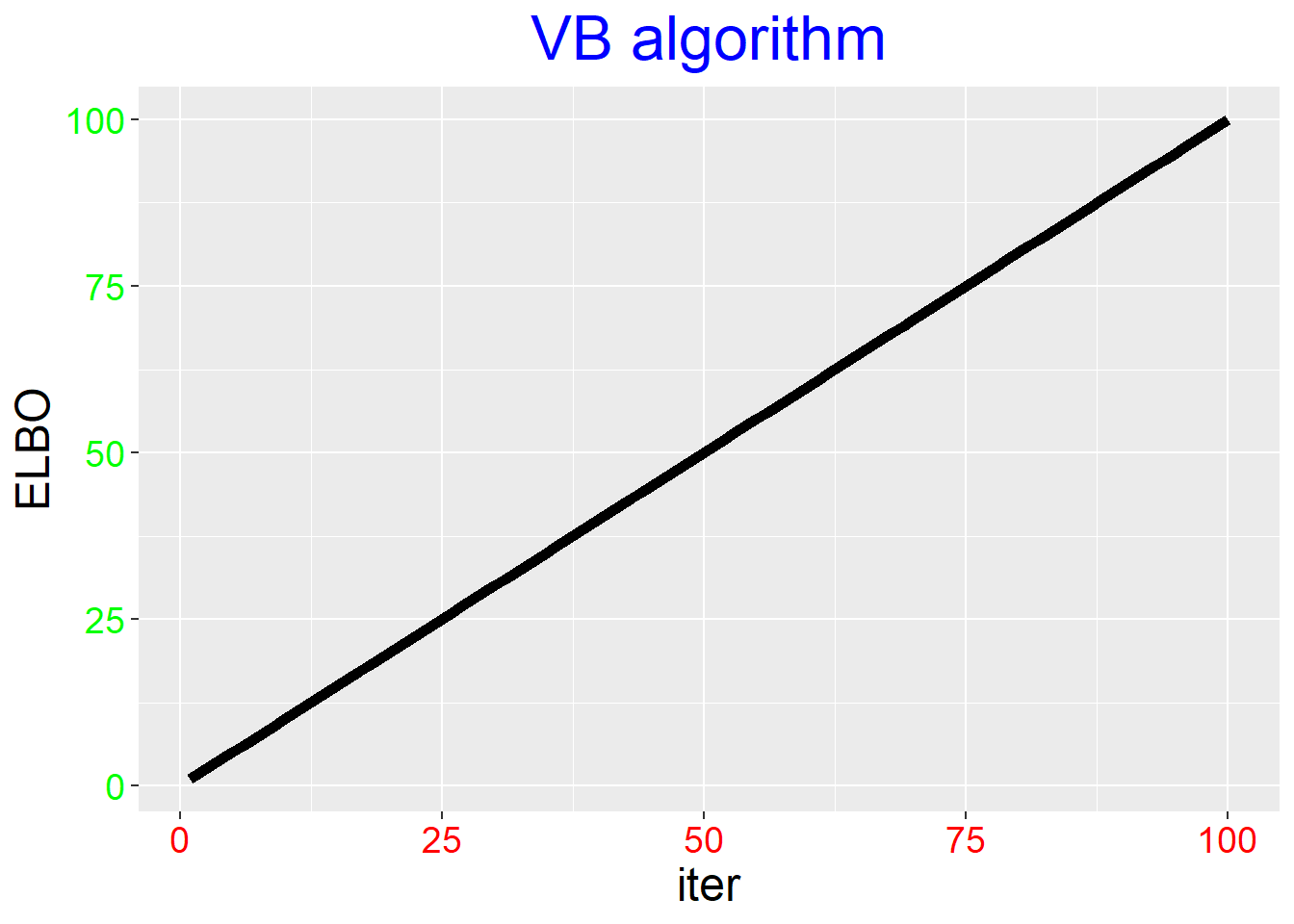
single_line_Plot <- function(y, x=NULL, x.name="iter", y.name="logLik",
title.name="Title",title.col="black", title.size=20, x.axis.col='black', x.axis.size=14, y.axis.col='black', y.axis.size=14, xlab.size=18, ylab.size=18, line.size=2){
library(ggplot2)
if(is.null(x)) x <- 1:length(y)
if(length(x) != length(y)) stop("x and y must have same length!")
p1 <- ggplot(data=NULL,
aes(x=x, y=y))
p1 + geom_line(size=line.size) + ggtitle(title.name) + xlab(x.name) + ylab(y.name) +
theme(plot.title = element_text(hjust = 0.5),
axis.text.x=element_text(size=x.axis.size,color=x.axis.col),
axis.text.y=element_text(size=y.axis.size, color=y.axis.col),
axis.title.x = element_text(size=xlab.size, color='black'),
axis.title.y = element_text(size=ylab.size, color='black'),
title= element_text(size=title.size, color=title.col)
)
}
single_line_Plot(1:10)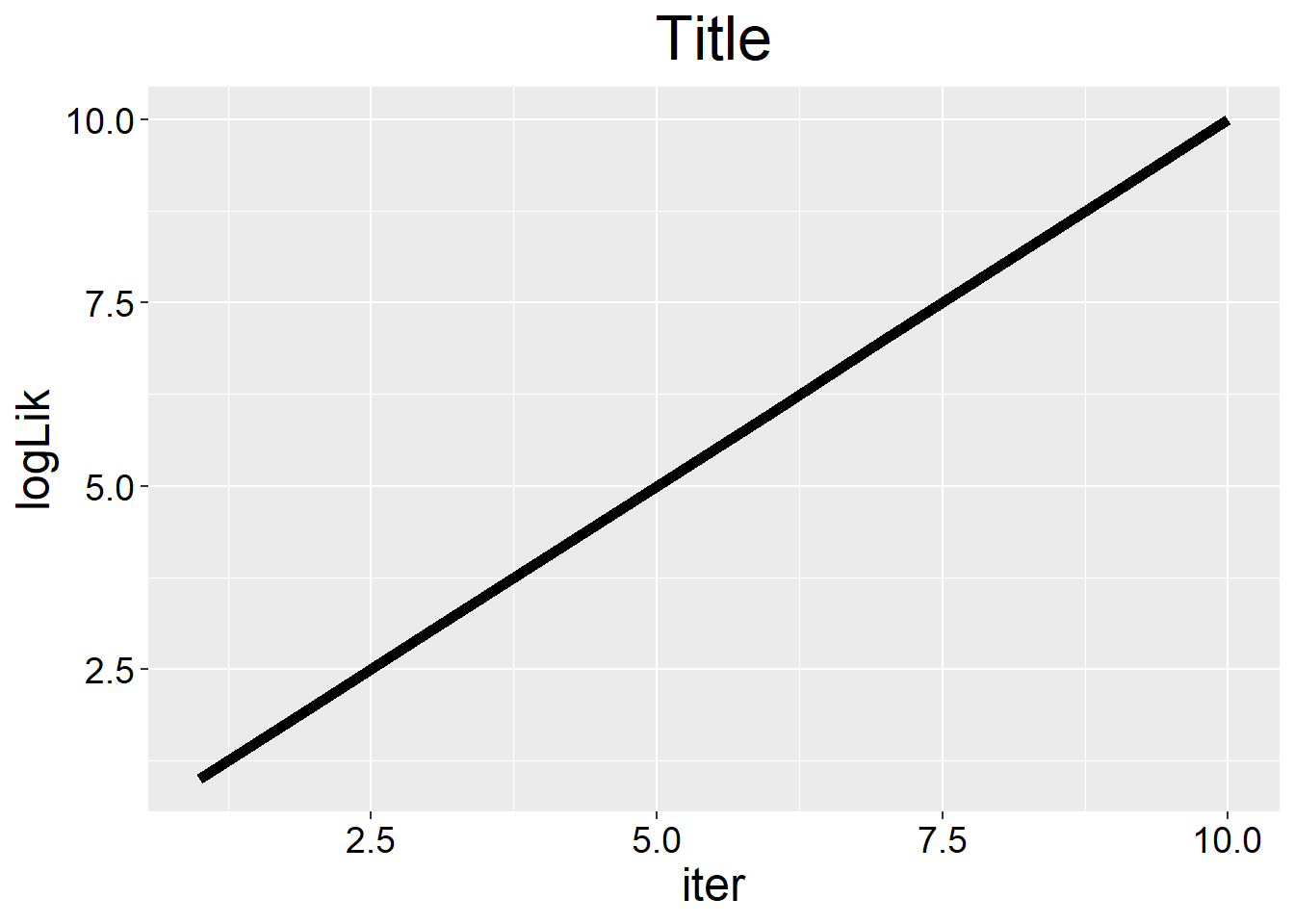
↑ 返回目录页
### Work on scale of axis{#yscale}
simutool::boxPlot(y=timeMat, title.name = " ",y.name = 'Time(sec.)') + scale_y_log10()Work on layout
类似par(mfrow=c(2,2))的用法包:
library(grid)
grid.newpage()
pushViewport(viewport(layout = grid.layout(2,2)))
print(p1, vp = viewport(layout.pos.row = 1, layout.pos.col = 1))
print(p2, vp = viewport(layout.pos.row = 1, layout.pos.col = 2))
print(p3, vp = viewport(layout.pos.row = 2, layout.pos.col = 1))
print(p4, vp = viewport(layout.pos.row = 2, layout.pos.col = 2))Or use the patchwork package
library(patchwork)
p <- p1+p2+p3+p4 +
plot_layout(byrow = T, nrow=2, ncol=2)Work on colors
cols <- c("antiquewhite2", "dodgerblue2","palegreen2","goldenrod2","red",
"antiquewhite4","dodgerblue4","palegreen4","goldenrod4",
"blue4", "brown")
# check the color codes
library(colorspace)
ramp.list = adjust_transparency(cols, alpha = 0.6) # Check the color codes in the specified transparency.
ramp.list
# plot the color bar to check the colors
barplot(rep(1, length(ramp.list)), axes = FALSE, space = 0, col = ramp.list)
cols[-5] <- ramp.list[-5] # save the colors we required.
manul_fill_col <- scale_fill_manual(values = cols)
# Then plot
p1 + geom_violin(trim=FALSE, scale = "width" )+
geom_boxplot(width=0.1, fill="white") + facet_wrap(PCs~Error,scales= "free" ) + labs(x=NULL)+
scale_x_discrete(breaks = NULL) + manul_fill_col +
thmem_used ggsci is a wonderful colors platte package, which can help you choose color automatically. ggcsi配色网址: http://www.sci666.net/60850.html
library(ggsci)
library(patchwork)
g1 <- clusterPlot(dlpfc2, label=clusters[-na_id], palette=NULL, size=0.05)
p1 <- g1 + scale_fill_jco() + labs(title= 'JCO') + guides(fill=F)
p2 <- g1 + scale_fill_npg() + labs(title= 'Nature')
p3 <- g1 + scale_fill_aaas()+ labs(title= 'Science')
p4 <- g1 + scale_fill_nejm()+ labs(title= 'NEJM')
p1 + p2 + p3 + p4
p5 <- g1 + scale_fill_lancet() + labs(title= 'Lancet')
p6 <- g1 + scale_fill_jama() + labs(title= 'JAMA')
p7 <- g1 + scale_fill_d3() + labs(title= 'd3')
p8 <- g1 + scale_fill_ucscgb()+ labs(title= 'UCSCGB')
p5 + p6 + p7 + p8
p5 <- g1 + scale_fill_locuszoom() + labs(title= 'Locuszoom')
p6 <- g1 + scale_fill_igv() + labs(title= 'IGV')
p7 <- g1 + scale_fill_startrek() + labs(title= 'Startrek')
p8 <- g1 + scale_fill_uchicago()+ labs(title= 'Uchicago')
p5 + p6 + p7 + p8
Auto save many figures
Ordinary plot in R
# filepath <- 'IncreasedModellingFiles/Model6_HuBei_Guaipoints/'
for(k in 1:n.HuBei){
# jpeg(file = paste0(filepath, '/', row.names(Zkt.HuBei)[k], '.jpg')) # 保存为jpg
pdf(file = paste0(filepath, '/', row.names(Zkt.HuBei)[k], '.pdf')) #保存为pdf
plot(cum_quezhen_pred[k, ], axes = F ,type='l', main=row.names(Zkt.HuBei)[k],
ylab='cum. identified illness', xlab='Date',col=2, lty=2)
lines(cum_quezhen[k,], col=1)
points(cum_quezhen[k,], col=1)
v1_tmp <- which(colnames(Zkt_pred) == guaipoint_vec[k])
abline(v=v1_tmp, col = 'blue')
text(x=v1_tmp, y=cum_quezhen_pred[k, round(TT.HuBei/2)],
guaipoint_vec[k])
axis(1, 1:ncol(Zkt_pred),labels= colnames(Zkt_pred))
axis(2)
grid()
dev.off()
} ggplot2:
filepath <- "D:\\LearnFiles\\Research paper\\ProPCA\\RealData"
n_ID <- length(name_ID12)
for(j in 1: n_ID){
cat('brain ', j, '\n')
clusters <- list()
clusters[[1]] <- clusterList12[[j]]$SSDRcluster
clusters[[2]] <- clusterList12[[j]][[paste0('BayesSpace', clustNumMat[j,3])]]
clusters[[3]] <- clusterList12[[j]]$GMRF
dlpfc <- readRDS(paste0("humanbrain\\", name_ID12[j],".rds") )
clusters[[4]] <- dlpfc@colData$layer_guess_reordered
na_id <- which(is.na(clusters[[4]]))
names(clusters) <- c("SSDRcluster", 'BayesSpace', "GMRF", "Groundtruth")
library(BayesSpace)
library(ggplot2)
## Plot the figures we require to save.
dlpfc2 <- dlpfc[,-na_id] # dropout the missing values
p1 <- clusterPlot(dlpfc2, label=clusters[[4]][-na_id], palette=NULL, size=0.05) +
scale_fill_viridis_d(option = "A", labels = 1:clustNumMat[j,1]) +
labs(title="groundtruth")
p2 <- clusterPlot(dlpfc2, label= as.factor(clusters[["SSDRcluster"]][-na_id]), palette=NULL, size=0.05) +
scale_fill_viridis_d(option = "A", labels = 1:clustNumMat[j,2]) +
labs(title=paste0("SSDRcluster: ARI=", round(AriMat[j,1],2)))
p3 <- clusterPlot(dlpfc2, label= as.factor(clusters[["BayesSpace"]][-na_id]), palette=NULL, size=0.05) +
scale_fill_viridis_d(option = "A", labels = 1:clustNumMat[j,3]) +
labs(title=paste0("BayesSpace",clustNumMat[j,3],": ARI=", round(AriMat[j,2],2)))
p4 <- clusterPlot(dlpfc2, label= as.factor(clusters[["GMRF"]][-na_id]), palette=NULL, size=0.05) +
scale_fill_viridis_d(option = "A", labels = 1:clustNumMat[j,4]) +
labs(title=paste0("GMRF: ARI=", round(AriMat[j,3],2)))
# pdf(file =) #保存为pdf
# layout the multiple figures
library(patchwork)
p <- p1+p2+p3+p4
# Start to save figures.
ggsave(filename= paste0(filepath, '\\Figures\\brain', name_ID12[j], 'method3.pdf'),
plot = p,width = 10, height = 8, units = "in", dpi = 1000)
}Work on theme
library(ggtext)
library(rlang)
library(ggthemes)
library(ggplot2)
element_textbox_highlight <- function(..., hi.labels = NULL, hi.fill = NULL,
hi.col = NULL, hi.box.col = NULL, hi.family = NULL) {
structure(
c(element_textbox(...),
list(hi.labels = hi.labels, hi.fill = hi.fill, hi.col = hi.col, hi.box.col = hi.box.col, hi.family = hi.family)
),
class = c("element_textbox_highlight", "element_textbox", "element_text", "element")
)
}
strip.text1 <- element_textbox_highlight(
family = NULL, size = 12, face = "bold",
fill = "white", box.color = "chartreuse4", color = "black",
halign = .5, linetype = 1, r = unit(5, "pt"), width = unit(1, "npc"),
padding = margin(5, 0, 3, 0), margin = margin(0, 1, 3, 1),
hi.labels = c("1997", "1998", "1999", "2000"),
hi.fill = "chartreuse4", hi.box.col = "black", hi.col = "white"
)
# This theme's background is filled with white, boarder with black.
thmem_used <- theme(axis.text.x=element_blank(),
axis.text.y=element_text(size=12, color=1),
axis.title.x = element_text(size=12, color='black'),
axis.title.y = element_text(size=14, color='black'),
strip.text = strip.text1,#element_text(size=12),
legend.direction = "horizontal", legend.position = "bottom",
legend.text=element_text(size=15),
panel.background= element_rect(fill = 'white', colour = 'black'))