R语言生信笔记
Table of Content
检查R包的下载次数:
For the DR.SC package:
Downloads each month (https://cranlogs.r-pkg.org/badges/DR.SC)
Downloads in last day (https://cranlogs.r-pkg.org/badges/last-day/DR.SC)
Grand total (https://cranlogs.r-pkg.org/badges/grand-total/DR.SC)
将R中的BLAS替换成OpenBLAS加快计算速度:
在64位windows下安装OpenBLAS的步骤如下: 从压缩文件OpenBLAS-v0.2.8-x86_64-Win.zip中解压libopenblas.dll到R目录下64目录中。将此目录中Rblas.dll文件名改成Rblas.dll.back作为备份。将libopenblas.dll文件名改成Rblas.dll。 下载网址:https://github.com/xianyi/OpenBLAS/releases
R向xlsx中输出多个sheets
直接稍微修改下面的code即可,方便快捷。注意输入sheet前数据务必先转化为data frame格式。
for(i in 1:9){
dat <- matrix(rnorm(50*4), 50, 4)
dat <- as.data.frame(dat)
colnames(dat) <- c('t value',' Pr(>|t|)',' FDR',' avg_log2FC')
# xlsx::write.xlsx(dat, file='DEGs.xlsx', sheetName = paste0('sheet', i), append = T)
}快速选择前n个差异表达基因的信息并画HeatMap
## DEG analysis based on the annotated labels
# install.packages('DR.SC')
suppressPackageStartupMessages(library(DR.SC) )## Warning: package 'spatstat.geom' was built under R version 4.0.5## Warning: package 'spatstat.data' was built under R version 4.0.4 seu <- gendata_RNAExp(height=20, width=20,p=200, K=4)
library(Seurat)## Attaching SeuratObject # standard log-normalization
seu <- NormalizeData(seu)
seu <- ScaleData(seu)## Centering and scaling data matrix Idents(seu) <- factor(seu$true_clusters)
dat <- FindAllMarkers(seu)## Calculating cluster 1## Calculating cluster 2## Calculating cluster 3## Calculating cluster 4 suppressPackageStartupMessages(library(dplyr) )## Warning: package 'dplyr' was built under R version 4.0.5 dat %>%
group_by(cluster) %>%
top_n(n = 10, wt = avg_log2FC) -> top10
seus <- subset(seu, downsample = 300)
## HeatMap
library(ggplot2)
p <- DoHeatmap(seus, features = top10$gene, label = F) +
theme(legend.text = element_text(size=16),
legend.title = element_text(size=18, face='bold'),
axis.text.y = element_text(size=14)) +
guides(shape = guide_legend(override.aes = list(size=4)))#+ NoLegend()
p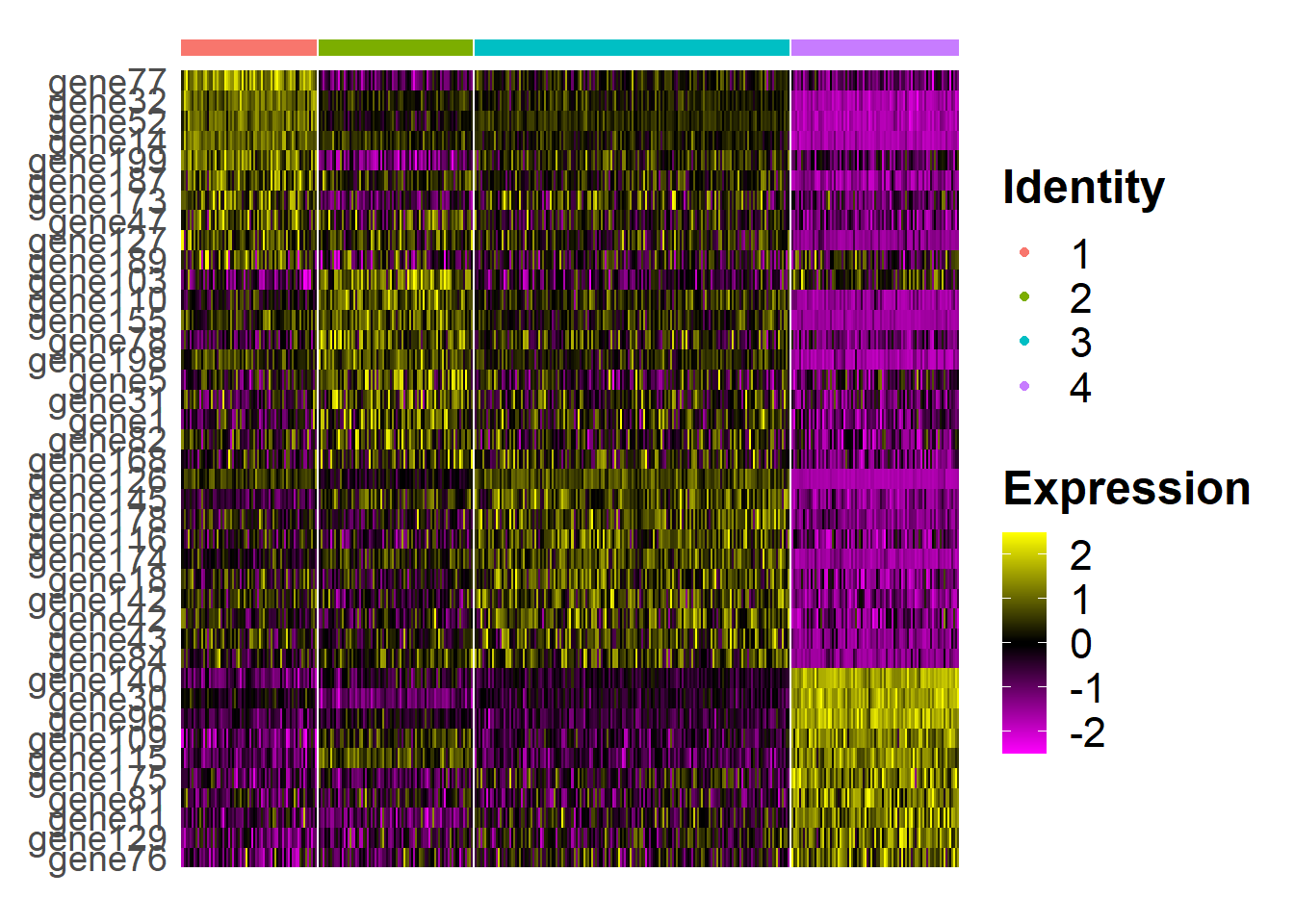
# ggsave(filename = 'heatMap_Brain1.pdf', plot = p,
# width = 12.5, height = 7, units = "in", dpi = 1000)基因symbols和ENSEMBL等ID的相互转化
## 1. transfor Human gene symbols to Ensembl
geneset <- c('AQP4', 'HPCAL1', 'FREM3', 'TRABD2A', 'KRT17',
'FABP7' , 'ADCYAP1' ,'PVALB',
'CCK' ,'ENC1','CUX2','RORB','NTNG2','MBP','PCP4',"SNAP25", "MOBP")
suppressPackageStartupMessages(library("org.Hs.eg.db") )## Warning: package 'BiocGenerics' was built under R version 4.0.5# remember to install it if you don't have it already
Ensembl <- mapIds(org.Hs.eg.db, keys = geneset,
keytype = "SYMBOL", column="ENSEMBL")## 'select()' returned 1:many mapping between keys and columnscat(Ensembl[1:4], '\n')## ENSG00000171885 ENSG00000115756 ENSG00000183090 ENSG00000186854# 2. transfor Human gene Ensembl to symbols
Symbol <- mapIds(org.Hs.eg.db, keys = Ensembl,
keytype = "ENSEMBL", column="SYMBOL")## 'select()' returned 1:many mapping between keys and columnscat(Symbol[1:4], '\n')## AQP4 HPCAL1 FREM3 TRABD2A